

I am a PhD student being advised by Luca Pinello at Harvard University Bioinformatics & Integrative Genomics Program and Massachusetts General Hospital.
My research interest lies in developing computational methods to better study the gene regulatory relationship. I work with single cell genomics and CRISPR screen data to study the relationship between genomic features and variant functions.
CRISPR-Cas system is a powerful toolkit to directly test the variant effect to various phenotypes. I am actively developing a Bayesian inference method using probabilistic programming language for CRISPR base-editor screen analysis that takes variable editing efficiency into account.
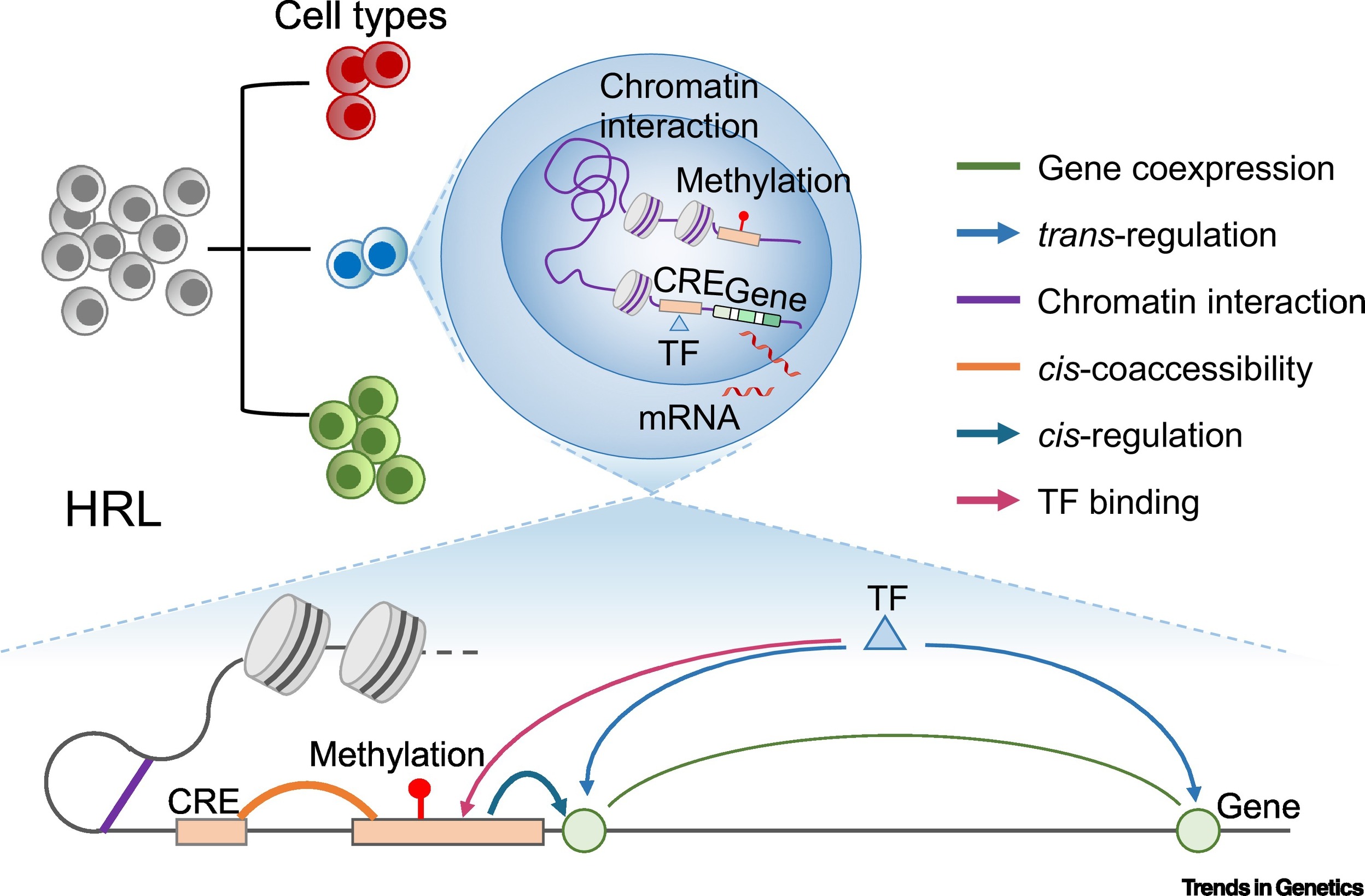
Uni and multi-modal single cell profile allows for studying gene regulatory relationship within and between modality. (Image: Li et al., 2020)

CRISPR genome editing technology allows for direct perturbation of genetic variants.
Another axis of my research interest is to build interpretable statistical methods that harmonize multimodal and multi-level biological knowledge utilizing graph representation learning.
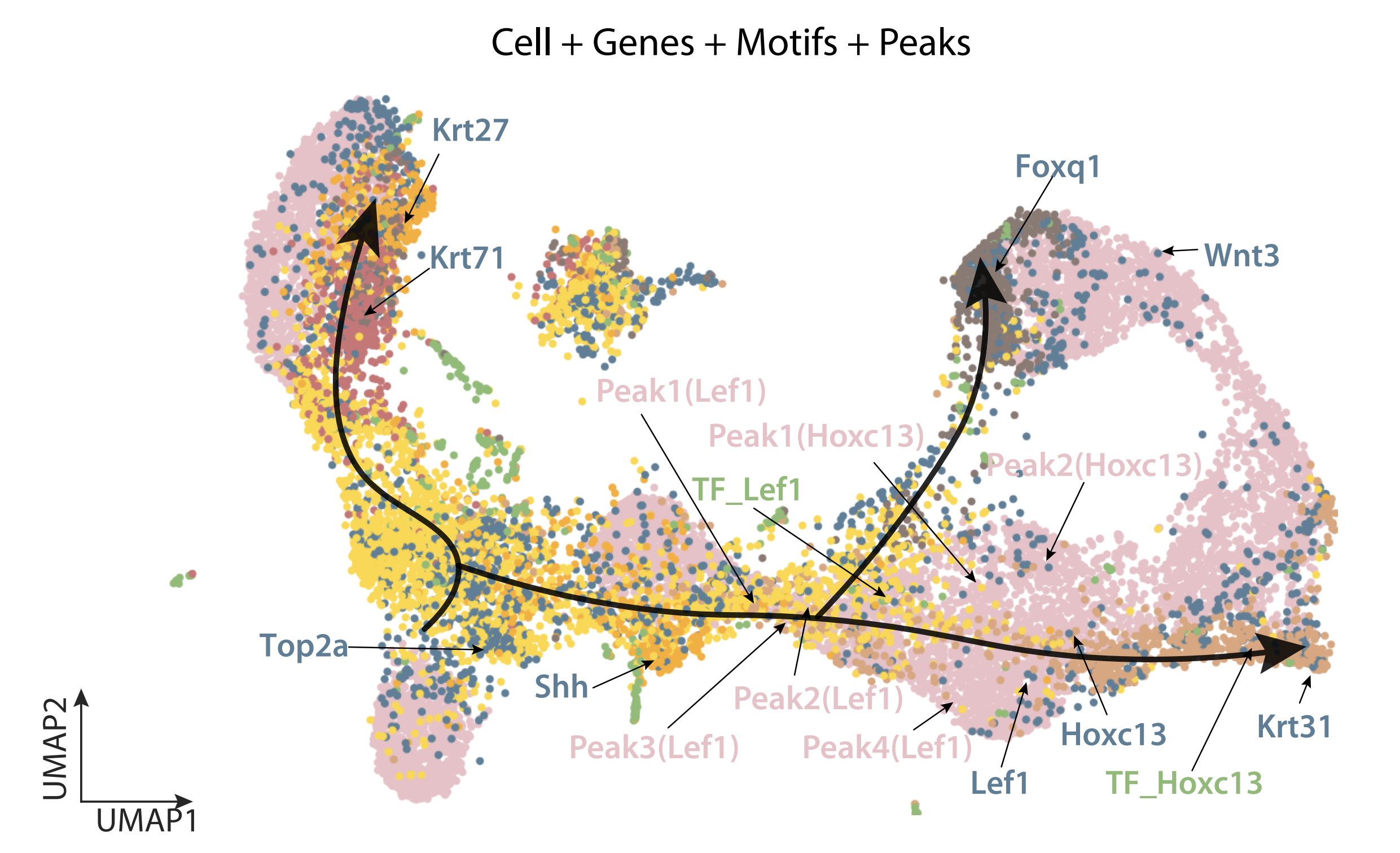
SIMBA, a graph representation learning based single cell embedding method allows joint analysis of cells, genes, motifs and peaks.